Team members, Eva, Manon, Miriam, and Charlotte, showcased their groundbreaking work through oral presentations, while Eline and Lieselot presented their research via an engaging poster session. |
| Elfride has received the 2023 Neurovision prize, awarded yearly by the Fondation JED Belgique. This is the fourth time Elfride is one of the prestigious Neurovision laureates. The jury was impressed by Elfride's work on "shedding light on the dark matter of the retinal genome, using a multi-omics approach". |
From 17-20th of October, 4 members of the De Baere Lab team attended the EMBO workshop on enhanceropathies: understanding enhancer function to understand human diseases, in Marseille (France). Lieselot presented a poster on one of the causes of North Carolina Macular Dystrophy (NCMD), namely non-coding duplications near IRX1. Chromatin interaction profiling of this region uncovered 10 candidate cis-regulatory elements (cCREs). Her poster details how she will investigate the underlying disease mechanisms of NCMD, a retinal enhanceropathy.
Charlotte presented a poster on her research deciphering the cis-regulatory region of FOXL2, a key factor in ovarian development. By combining her self-generated Hi-C and UMI-4C data in granulosa cells with publicly available OMICS data, she was able to identify 35 cCREs and 2 lncRNAs that will be validated using CRISPRi.
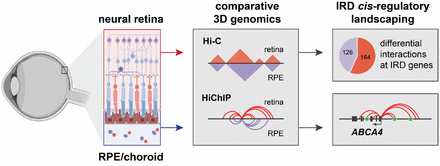
Eva presented a poster on a comparative 3D genome analysis between neural retina and RPE and the implications for the regulation of retinal disease genes. Lastly, also Victor presented a poster where he unravels the cis-regulatory landscape of ABCA4, in adult human retina.
Charlotte presented a poster on her research deciphering the cis-regulatory region of FOXL2, a key factor in ovarian development. By combining her self-generated Hi-C and UMI-4C data in granulosa cells with publicly available OMICS data, she was able to identify 35 cCREs and 2 lncRNAs that will be validated using CRISPRi.
Eva presented a poster on a comparative 3D genome analysis between neural retina and RPE and the implications for the regulation of retinal disease genes. Lastly, also Victor presented a poster where he unravels the cis-regulatory landscape of ABCA4, in adult human retina.
NEW PAPER ALERT!
By Alfredo Duenas Rey & Victor Lopez Soriano
Cross-species genome comparisons have revealed a substantial number of ultraconserved non-coding elements (UCNEs). Several of these elements have proved to be essential tissue- and cell type-specific cis-regulators of developmental gene expression. Here, we characterized a set of UCNEs as candidate CREs (cCREs) during retinal development and evaluated the contribution of their genomic variation to rare eye diseases, for which pathogenic non-coding variants are emerging. Integration of bulk and single-cell retinal multi-omics data revealed 594 genes under potential cis-regulatory control of UCNEs, of which 45 are implicated in rare eye disease. Mining of candidate cis-regulatory UCNEs in WGS data derived from the rare eye disease cohort of Genomics England revealed 178 ultrarare variants within 84 UCNEs associated with 29 disease genes. Overall, we provide a comprehensive annotation of ultraconserved non-coding regions acting as cCREs during retinal development which can be targets of non-coding variation underlying rare eye diseases.
By Alfredo Duenas Rey & Victor Lopez Soriano
Cross-species genome comparisons have revealed a substantial number of ultraconserved non-coding elements (UCNEs). Several of these elements have proved to be essential tissue- and cell type-specific cis-regulators of developmental gene expression. Here, we characterized a set of UCNEs as candidate CREs (cCREs) during retinal development and evaluated the contribution of their genomic variation to rare eye diseases, for which pathogenic non-coding variants are emerging. Integration of bulk and single-cell retinal multi-omics data revealed 594 genes under potential cis-regulatory control of UCNEs, of which 45 are implicated in rare eye disease. Mining of candidate cis-regulatory UCNEs in WGS data derived from the rare eye disease cohort of Genomics England revealed 178 ultrarare variants within 84 UCNEs associated with 29 disease genes. Overall, we provide a comprehensive annotation of ultraconserved non-coding regions acting as cCREs during retinal development which can be targets of non-coding variation underlying rare eye diseases.
Prof. Elfride De Baere, received a generous fund granted by the King Baudouin Foundation last week. This grant will be used for further research studies in our research group on functional genomics in the retina.
Dr. Anouk Georges (MD, PhD) joined us for a very interesting seminar about comparing the transcriptome of developing native and iPS-derived mouse retinae by scRNA-seq & ATAC-seq.
NEW PAPER ALERT!
By Alexandre Segers
RNA-sequencing (RNA-seq) is increasingly used to diagnose patients with rare diseases by prioritising genes with aberrant expression and/or splicing. State-of-the-art methods for detecting aberrant expression and splicing, however, are extremely slow. The latter, also discard much information because they only use junction reads to infer aberrant splicing. In this contribution, we show that replacing the offset for library size unlocks conventional bulk RNA-seq workflows for fast and scalable differential usage, aberrant splicing and expression analyses. Our method, saseR, is several orders of magnitude faster than the state-of-the-art methods and dramatically outperforms these in terms of sensitivity and specificity for aberrant splicing, while being on par with these inferring differential usage and aberrant expression. Finally, our framework is also very flexible and can be used for all applications that involve the analysis of proportions of short- or long RNA-seq read counts.
By Alexandre Segers
RNA-sequencing (RNA-seq) is increasingly used to diagnose patients with rare diseases by prioritising genes with aberrant expression and/or splicing. State-of-the-art methods for detecting aberrant expression and splicing, however, are extremely slow. The latter, also discard much information because they only use junction reads to infer aberrant splicing. In this contribution, we show that replacing the offset for library size unlocks conventional bulk RNA-seq workflows for fast and scalable differential usage, aberrant splicing and expression analyses. Our method, saseR, is several orders of magnitude faster than the state-of-the-art methods and dramatically outperforms these in terms of sensitivity and specificity for aberrant splicing, while being on par with these inferring differential usage and aberrant expression. Finally, our framework is also very flexible and can be used for all applications that involve the analysis of proportions of short- or long RNA-seq read counts.
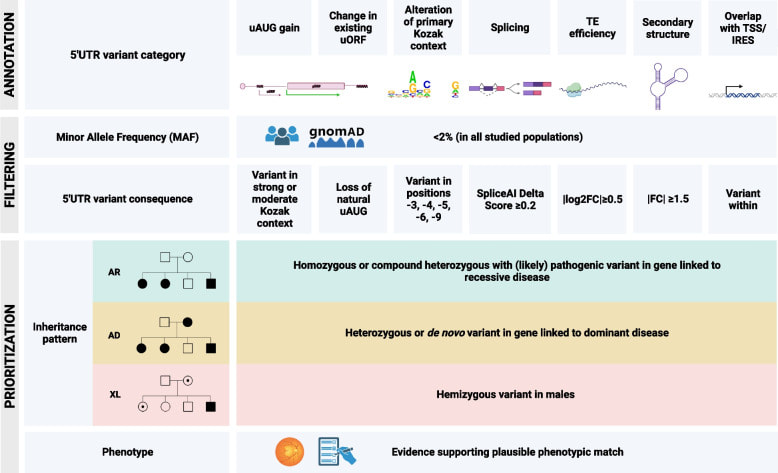
| NEW PAPER ALERT! By Alfredo Duenas Rey This study focuses on the significance of 5' untranslated region (5'UTR) variants in inherited retinal diseases (IRDs). It presents a comprehensive approach involving the analysis of retinal RNA data for 378 IRD genes, assessment of 5'UTR coverage in whole exome sequencing data, and evaluation of 5'UTR variants in large patient cohorts. The study identified 11 potentially pathogenic variants in 10 genes, with two of them validated as having functional effects. Notably, the MERTK:c.-125G>A variant reduced luciferase mRNA levels and activity, while the RDH12:c.-123C>T variant, found in conjunction with another reported variant, resulted in reduced RDH12 protein levels. This research underscores the importance of 5'UTR variants in IRDs and offers a systematic strategy for annotating and validating such variants, applicable to other inherited diseases. |
| NEW PAPER ALERT! By Dr. Eva D'haene & Victor Lopez Soriano Through extensive comparative 3D genome mapping, based on genome-wide (Hi-C), promoter-centric (HiChIP) and locus-specific (UMI-4C) assays of human neural retina and RPE, we have shown that gene regulation at key IRD loci is likely mediated by tissue-specific chromatin interactions. These findings do not only provide insight into tissue-specific regulatory landscapes of IRD genes, but also delineate the search space for non-coding genomic variation underlying unsolved IRD. |
| GREAT NEWS! ProgRET will create a multidisciplinary and intersectoral European training network focusing on the mechanisms, diagnosis and therapy of dominantly inherited retinal diseases (IRD). IRD represent a major cause of blindness, affecting 350,000 people in Europe. IRD have long been considered incurable, however major advances have led to groundbreaking new treatments. Today, the most important challenges in the IRD field relate to an unsolved genetic diagnosis, unknown disease mechanisms and gene therapy development for autosomal dominant IRD (adIRD), representing 25%–40% of all IRD cases. We have demonstrated an emerging role for splicing factors, structural variants and non-coding defects in patients with adIRD, and developed novel disease models and gene therapies for adIRD. ProgRET aims to dissect adIRD mechanisms using retinal stem cell and aquatic animal models, to advance adIRD diagnostics using a single-molecule multi-omics framework, and to develop innovative treatments based on RNA therapy and CRISPR-genome editing. These challenges will be tackled by integrating unique expertise and cutting-edge technology within ProgRET, including (multi-)omics, bioinformatics, functional genomics, RNA biology, gene regulation, stem cell technology, retinal organoids, animal models, genome editing and gene therapy. ProgRET will give Doctoral Candidates (DCs) unparalleled training opportunities in outstanding academic and industrial settings through training-by-research via individual research projects, secondments, and network-wide training sessions. All individual training and research activities will provide each DC with the necessary skills in academic and industrial research. ProgRET will make a career in both sectors attractive and improve their career prospects. Finally, our multidisciplinary network offers a unique opportunity to accelerate the understanding, diagnostics and therapeutics for adIRD in Europe, and to translate research findings to healthcare and society. |
Three fabulous keynote speakers were invited to the RARE-MED symposium covering non-coding variantion, Nicky Whiffin (Oxford Universoty), functional genomics, Musa Mhlanga (Radboud University, Niijmegen, The Netherlands) and RNA therapy of Neurological Diseases, Willeke Van Roon-Mom (leiden University Medical Center & Dutch center for RNA therapeutics, Leiden, The Netherlands). Apart from this, several members of our team presented their research (posters and oral presentations). Special acknowledgement to our team member Eline Van Vooren who won the price for Best Poster with her research on RPE65 VUS assessment.
We are looking forward to next year's edition!
We are looking forward to next year's edition!
No less than six lab members had an oral talk at the 2022 ESHG meeting. Sarah Vergult and Eva D'Haene presented on Structural variants disrupt a critical regulatory region downstream of FOXG1 and Mapping the 3D genome of the human retina and its role in retinal disease, respectively. Stijn Vandesompele's talk was titled Dual molecular effects of constitutional SF3B2 variants cause a novel dominant spliceosomopathy displaying retinitis pigmentosa or developmental skeletal anomalies and Rocio discussed A novel neurodevelopmental syndrome caused by loss-of-function of the Zinc Finger Homeobox 3 (ZFHX3) gene. Victor López Soriano had a talk on An integrated approach using multi-omics data to dissect cis-regulatory role of ultraconserved non-coding elements (UCNRE) in human retina in the same session as Alfredo, who spoke Identification and charaterization of a novel retina-specific lncRNA upstream ABCA4 with a potential role in ABCA4-associated inherited retinal disease. Congrats to all on the very nice presentations! Miriam Bauwens had a poster presentation.
Three Lab De Baere members had an oral presentation and six had a poster presentation at the 22th Belgian Society for Human Genetics meeting in Bruges. Congrats to Eva, Burcu and Rocio for their oral presentations on Mapping the 3D genome of the human retina and its role in retinal disease, Single-cell transcriptional dynamics and in vivo enhancer assays provide insight into gene regulatory networks of PRDM13 and IRX1 implicated in North Carolina macular dystrophy and A novel neurodevelopmental syndrome caused by loss-of-function of the Zinc Finger Homeobox 3 (ZFHX3) gene, respectively. Posters by Miriam, Marta, Alfredo, Charlotte, Marjolein, Victor were visable during the poster sessions. We also had fun during the conference dinner, party and especially during the suprise act by science comedian Lieven Scheiren.