The European Vision Institute has announced its list of top women in European Vision Research and ophthalmology for 2021. Elfride De Baere was granted a place on the list. Congratulations Elfride
Münevver BurcU Cicekdal won the poster prize at the 18th international xenopus conference 202130/8/2021
Rare-med walks, runs, bikes to support rare diseases! Support our research on rare diseases!22/2/2021
We would like to invite you to the online kick-off symposium of the RARE-MED consortium on November 27, 2020, from 14-17.30 p.m. Registration is free of charge and can be done here. Please register if you wish to attend, as only registered people will receive a link for the symposium. The symposium program is displayed below.
RARE-MED is a multidisciplinary UGent consortium for basic and translational research on precision medicine for rare diseases, to address missing heritability using systems genetics and functional genomics, to facilitate disease modelling using CRISPR/Cas9-mediated genome editing of aquatic model organisms (zebrafish, Xenopus) and of cellular systems, to introduce new gene therapies based on antisense oligonucleotide- or CRISPR/Cas9-based genome editing. In the context of this 21ZAP consortium, 3 research professors were appointed: Prof. dr. Sarah Vergult, prof. dr. Kris Vleminckx, and prof. dr. Frauke Coppieters.
Giulia Ascari virtually defended her PhD thesis on June 18 2020. Congratulations to our doctor, whose favorite genes are RCBTB1 & CEP78.
On the 27th of April 2020, Marlies Saelaert virtually defended her PhD thesis. This was an unforgettable experience. Congratulations!
Our new team member, Karolien Aelbrecht, virtually defended successfully her PhD thesis on April 28 2020. Well done, congratulations Karolien!
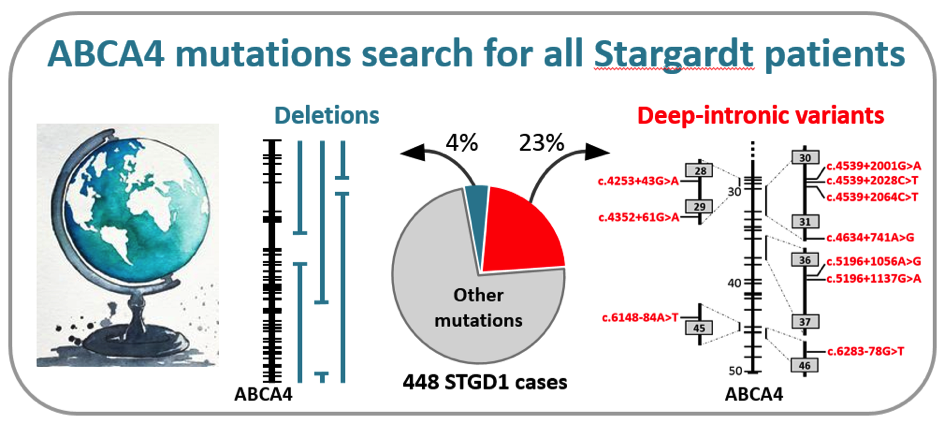
In a recently published manuscript in the prestigious journal Genetics in Medicine, a large group of collaborators, led by dr. Claire-Marie Dhaenens and prof. Frans Cremers in the Department of Human Genetics in Nijmegen, the Netherlands, identified the causal mutations in the ABCA4 gene in 448 individuals with Stargardt disease (STGD1). They employed a cost-effective method, based on so-called smMIPs, to sequence the complete ABCA4 gene consisting of 128,313 base pairs. Through a semi-automated procedure, they tested more than 1,000 probands with STGD1 and allied maculopathies for the presence of causal mutations (Khan et al. Genet Med, in press).
Remarkably, they found 105 causal mutations residing in the introns of the ABCA4 gene that led to the disruptive insertion of these non-coding sequences in the messenger RNA and thus a non-functional ABCA4 protein. Among 13 novel RNA insertions identified in splicing assays, they found two intriguing complex RNA defects due to variants in introns 13 and 44. In addition, they found 16 novel large deletions, two of which were complex. Altogether, these ‘hidden mutations’ were found in 27% of the 448 genetically solved cases, illustrating the unusually high proportion of these types of mutations in STGD1. This study was made possible through the collaboration of 75 scientists and clinicians from 21 countries from all over the world. Due to the low sequencing costs (€ 30,- per case), the research team in Nijmegen now offers this test to any individual with STGD1 or allied maculopathy in the world, at no costs, on a research basis. This work was made possible through several grants, the most significant of which were from the RetinaUK, Fighting Blindness Ireland, Foundation Fighting Blindness USA, and the EU.
The John W. Mouton Pro Retina grant that was awarded to prof. Elfride De Baere for her research project Precision medicine in inherited blindness using integrated omics in patient-derived stem cell models was mentioned in the physicians' paper. To read the article click here.
Prof. De Baere was chosen as the Visionary for the first quarter of 2020 by vision-research.eu. To read more about her work on precision medicine to understand missing heritability in inherited retinal diseases, you can click here.
|
De Baere Lab © 2024