|
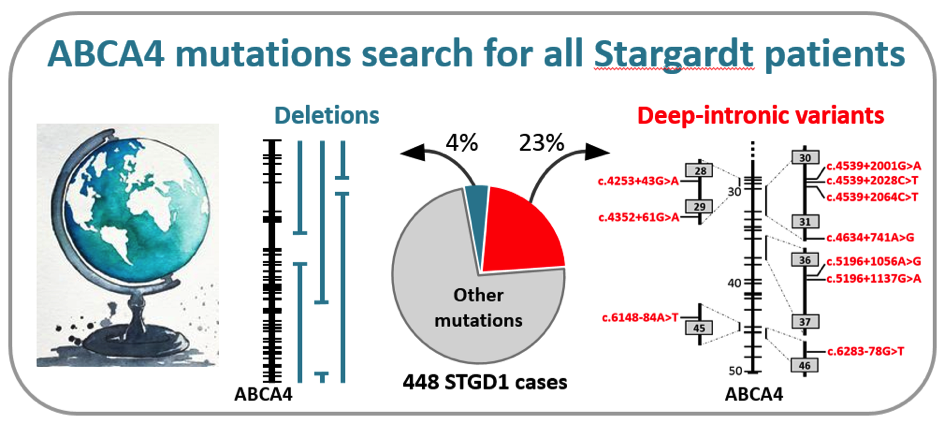
In a recently published manuscript in the prestigious journal Genetics in Medicine, a large group of collaborators, led by dr. Claire-Marie Dhaenens and prof. Frans Cremers in the Department of Human Genetics in Nijmegen, the Netherlands, identified the causal mutations in the ABCA4 gene in 448 individuals with Stargardt disease (STGD1). They employed a cost-effective method, based on so-called smMIPs, to sequence the complete ABCA4 gene consisting of 128,313 base pairs. Through a semi-automated procedure, they tested more than 1,000 probands with STGD1 and allied maculopathies for the presence of causal mutations (Khan et al. Genet Med, in press).
Remarkably, they found 105 causal mutations residing in the introns of the ABCA4 gene that led to the disruptive insertion of these non-coding sequences in the messenger RNA and thus a non-functional ABCA4 protein. Among 13 novel RNA insertions identified in splicing assays, they found two intriguing complex RNA defects due to variants in introns 13 and 44. In addition, they found 16 novel large deletions, two of which were complex. Altogether, these ‘hidden mutations’ were found in 27% of the 448 genetically solved cases, illustrating the unusually high proportion of these types of mutations in STGD1. This study was made possible through the collaboration of 75 scientists and clinicians from 21 countries from all over the world. Due to the low sequencing costs (€ 30,- per case), the research team in Nijmegen now offers this test to any individual with STGD1 or allied maculopathy in the world, at no costs, on a research basis. This work was made possible through several grants, the most significant of which were from the RetinaUK, Fighting Blindness Ireland, Foundation Fighting Blindness USA, and the EU. Comments are closed.
|
De Baere Lab © 2024