|
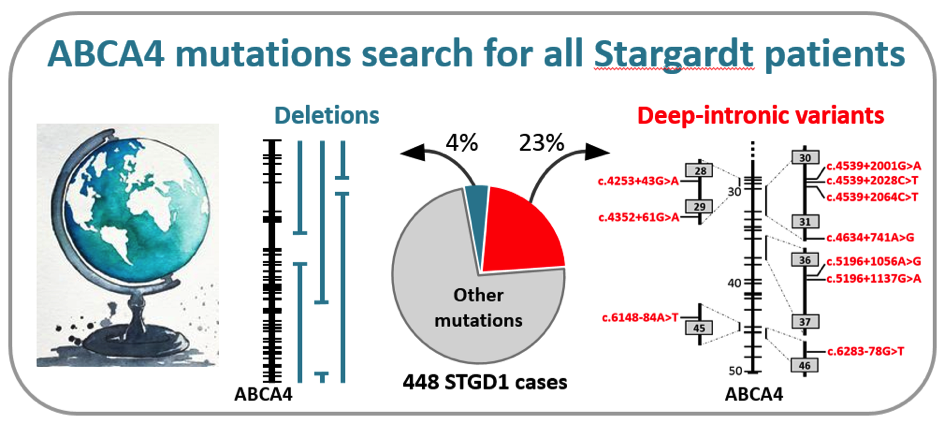
In a recently published manuscript in the prestigious journal Genetics in Medicine, a large group of collaborators, led by dr. Claire-Marie Dhaenens and prof. Frans Cremers in the Department of Human Genetics in Nijmegen, the Netherlands, identified the causal mutations in the ABCA4 gene in 448 individuals with Stargardt disease (STGD1). They employed a cost-effective method, based on so-called smMIPs, to sequence the complete ABCA4 gene consisting of 128,313 base pairs. Through a semi-automated procedure, they tested more than 1,000 probands with STGD1 and allied maculopathies for the presence of causal mutations (Khan et al. Genet Med, in press).
Remarkably, they found 105 causal mutations residing in the introns of the ABCA4 gene that led to the disruptive insertion of these non-coding sequences in the messenger RNA and thus a non-functional ABCA4 protein. Among 13 novel RNA insertions identified in splicing assays, they found two intriguing complex RNA defects due to variants in introns 13 and 44. In addition, they found 16 novel large deletions, two of which were complex. Altogether, these ‘hidden mutations’ were found in 27% of the 448 genetically solved cases, illustrating the unusually high proportion of these types of mutations in STGD1. This study was made possible through the collaboration of 75 scientists and clinicians from 21 countries from all over the world. Due to the low sequencing costs (€ 30,- per case), the research team in Nijmegen now offers this test to any individual with STGD1 or allied maculopathy in the world, at no costs, on a research basis. This work was made possible through several grants, the most significant of which were from the RetinaUK, Fighting Blindness Ireland, Foundation Fighting Blindness USA, and the EU.
The John W. Mouton Pro Retina grant that was awarded to prof. Elfride De Baere for her research project Precision medicine in inherited blindness using integrated omics in patient-derived stem cell models was mentioned in the physicians' paper. To read the article click here.
Prof. De Baere was chosen as the Visionary for the first quarter of 2020 by vision-research.eu. To read more about her work on precision medicine to understand missing heritability in inherited retinal diseases, you can click here.
Every month, ERN-EYE invites you to meet an active person within the network through a short interview. This month, it's prof. Elfride de Baere who accepted to answer their questions. You can read the entire interview here or watch the video below.
The 20th BeSHG meeting "Genome for all?" was held in Brussels on the 6th of March, 2020. We are very proud that Stijn won best oral presentation for his talk "Whole genome sequencing and 4C techniques provide novel insights into the genetic architecture and mechanisms underlying North Carolina macular dystrophy, a cis-regulatory disease". Congratulations Stijn!
Introduction
Every two years, the Fund John W. Mouton Pro Retina (managed by the King Baudouin Foundation) awards a grant of € 30.000 to a researcher working at a university or research center in Europe that is pursuing medical scientific research in the field of the pathologies of the retina. The fund is interested in all kind of retinal diseases (genetic, diabetic, hypertensive, post-prematurity, degenerative, toxic retinopathy, etc.) with exception of retinal tumors. The work may concern clinical research as well as basic research including a concrete perspective on further clinical work. In 2019, the John W. Mouton Pro Retina grant was awarded to prof. Elfride De Baere for her research project Precision medicine in inherited blindness using integrated omics in patient-derived stem cell models. Scientific abstract Inherited retinal disease (IRD) is a major cause of early-onset blindness affecting 2 million people worldwide. Significant advances have been made in the genomic underpinnings of IRD, which has culminated in novel therapies entering the clinic. Despite this progress, there are important knowledge gaps that hamper a molecular diagnosis in over half of the cases. We and others have shown a role for non-coding variation in undiagnosed IRD and demonstrated these are novel targets for therapy. Here, it is our main goal to accelerate diagnosis in unsolved IRD and to uncover novel targets for intervention. First, we will generate human cellular models of undiagnosed monoallelic patients with suspected recessive IRD. Second, we will establish an integrated omics framework to accelerate diagnosis in unsolved IRD. Third, we will translate research findings to the clinic through ERN-EYE and patient advocacy organizations. Our multidisciplinary approach involves ophthalmic genetics/genomics, (functional) genomics, bioinformatics, transcriptomics, proteomics, statistical genomics, and stem cell technology. Our expertise combined with a strong track record and international network offer a unique opportunity to address unmet needs and to pave the way to precision medicine in IRD. Lay summary Inherited retinal diseases represents major cause of certifiable blindness in the working age and childhood population. Despite a revolution in DNA technology that allowed to find genetic causes in half of the cases and that has culminated in successful gene therapies, essential knowledge is lacking to establish a genetic diagnosis in the remaining half of the patients, representing about 175.000 people in Europe. With this project we aim to improve the discovery of hidden mutations in inherited retinal disease, often residing in the ‘dark matter’ of the genome, and to find novel targets for intervention. From blood cells of patients with undiagnosed inherited retinal diseases we will make stem cells that we will let develop to cells that mimic light-sensitive retina cells (photoreceptors) that express disease-causing genes of interest. We will perform large-scale DNA, RNA, and protein studies on these cells from patients and controls. By integrating these three layers of information, we will develop a sophisticated strategy to pinpoint hidden mutations in undiagnosed patients. These mutations may represent novel targets for intervention studies. This proposal has expected health impact, accelerating a definite diagnosis in inherited retinal diseases, improving genetic counseling, reproductive options, disease prognosis and management and ultimately offering therapeutic opportunities. We will transfer results from our study to the patients through our local clinical and international networks. At the end of 2019, Sarah Naessens succesfully defended her PhD thesis on "Recurrent coding and rare non-coding targets for treatment in inherited retinal diseases". Congratulations Sarah and good luck with your future challenges!
ResearchUGent started off the RARE-MED consortium on precision medicine for rare diseases with 3 new research professors Sarah Vergult, Kris Vleminckx & Frauke Coppieters. Looking forward to addressing missing heritability, modeling disease, and introducing new gene therapies!
RARE-MED is a multidisciplinary consortium for basic and translational research on precision medicine for rare diseases, to address missing heritability using systems genetics and functional genomics, to facilitate disease modelling using CRISPR/Cas9-mediated genome editing of aquatic model organisms (zebrafish, Xenopus) and of cellular systems, to introduce new gene therapies based on antisense oligonucleotide- or CRISPR/Cas9-based genome editing. RARE-MED will bring UGent in a unique strategic position as an international reference center for rare disease
ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Bauwens M, Garanto A, Sangermano R, Naessens S, Weisschuh N, De Zaeytijd J, Khan M2, Sadler F5, Balikova I, Van Cauwenbergh C1, Rosseel T, Bauwens J, De Leeneer K, De Jaegere S, Van Laethem T, De Vries M, Carss K, Arno G, Fakin A, Webster AR, de Ravel de l'Argentière TJL, Sznajer Y, Vuylsteke M, Kohl S, Wissinger B, Cherry T, Collin RWJ, Cremers FPM, Leroy BP, De Baere E. Genet Med. (2019). You can read the abstract below; for the full text click here.
Purpose: ABCA4-associated disease, a recessive retinal dystrophy, is hallmarked by a large proportion of patients with only one pathogenic ABCA4 variant, suggestive for missing heritability. Methods: By locus-specific analysis of ABCA4, combined with extensive functional studies, we aimed to unravel the missing alleles in a cohort of 67 patients (p), with one (p = 64) or no (p = 3) identified coding pathogenic variants of ABCA4. Results: We identified eight pathogenic (deep-)intronic ABCA4 splice variants, of which five are novel and six structural variants, four of which are novel, including two duplications. Together, these variants account for the missing alleles in 40.3% of patients. Furthermore, two novel variants with a putative cis-regulatory effect were identified. The common hypomorphic variant c.5603A>T p.(Asn1868Ile) was found as a candidate second allele in 43.3% of patients. Overall, we have elucidated the missing heritability in 83.6% of our cohort. In addition, we successfully rescued three deep-intronic variants using antisense oligonucleotide (AON)-mediated treatment in HEK 293-T cells and in patient-derived fibroblast cells. Conclusion: Noncoding pathogenic variants, novel structural variants, and a common hypomorphic allele of the ABCA4 gene explain the majority of unsolved cases with ABCA4-associated disease, rendering this retinopathy a model for missing heritability in autosomal recessive disorders. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Sangermano R, Garanto A, Khan M, Runhart EH, Bauwens M, Bax NM, van den Born LI, Khan MI, Cornelis SS, Verheij JBGM, Pott JR, Thiadens AAHJ, Klaver CCW, Puech B, Meunier I2, Naessens S, Arno G, Fakin A, Carss KJ, Raymond FL, Webster AR, Dhaenens CM, Stöhr H, Grassmann F, Weber BHF, Hoyng CB, De Baere E, Albert S, Collin RWJ, Cremers FPM. Genet Med. (2019). You can read the abstract below; for the full text click here. Purpose: Using exome sequencing, the underlying variants in many persons with autosomal recessive diseases remain undetected. We explored autosomal recessive Stargardt disease (STGD1) as a model to identify the missing heritability. Methods: Sequencing of ABCA4 was performed in 8 STGD1 cases with one variant and p.Asn1868Ile in trans, 25 cases with one variant, and 3 cases with no ABCA4 variant. The effect of intronic variants was analyzed using in vitro splice assays in HEK293T cells and patient-derived fibroblasts. Antisense oligonucleotides were used to correct splice defects. Results: In 24 of the probands (67%), one known and five novel deep-intronic variants were found. The five novel variants resulted in messenger RNA pseudoexon inclusions, due to strengthening of cryptic splice sites or by disrupting a splicing silencer motif. Variant c.769-784C>T showed partial insertion of a pseudoexon and was found in cis with c.5603A>T (p.Asn1868Ile), so its causal role could not be fully established. Variant c.4253+43G>A resulted in partial skipping of exon 28. Remarkably, antisense oligonucleotides targeting the aberrant splice processes resulted in (partial) correction of all splicing defects. Conclusion: Our data demonstrate the importance of assessing noncoding variants in genetic diseases, and show the great potential of splice modulation therapy for deep-intronic variants.
|
De Baere Lab © 2024